Human, Mouse, and Rat Anti-Histone H1 Antibody Product Attributes
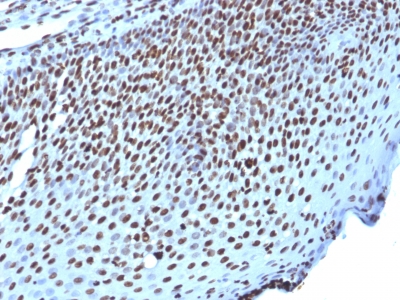
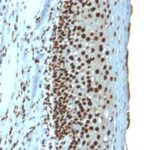
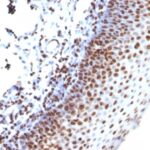
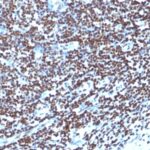
Histone H1 Previously Observed Antibody Staining Patterns
Observed Subcellular, Organelle Specific Staining Data:
Anti-H1F0 antibody staining is expected to be primarily localized to the nuclear bodies and nucleus. There is variability in either the signal strength or the localization of signal in nucleus from cell to cell.
| Histone H1 General Information | |
|---|---|
| Alternate Names | |
| Histone H1 | |
| Molecular Weight | |
| ~30kDa | |
| Chromosomal Location | |
| 3q21-q22; 6q21-q22; 22q13.1 | |
| Curated Database and Bioinformatic Data | |
| Gene Symbol | H1 |
| Entrez Gene ID | 3005 |
| Ensemble Gene ID | ENSG00000189060 |
| RefSeq Protein Accession(s) | NP_005309 |
| RefSeq mRNA Accession(s) | NM_005318 |
| RefSeq Genomic Accession(s) | NC_000022, NC_018933 |
| UniProt ID(s) | P07305 |
| UniGene ID(s) | P07305 |
| HGNC ID(s) | 4714 |
| Cosmic ID(s) | H1F0 |
| KEGG Gene ID(s) | hsa:3005 |
| PharmGKB ID(s) | PA29092 |
| General Description of Histone H1. | |
| Eukaryotic histones are basic, water-soluble nuclear proteins that form hetero-octameric nucleosome particles by wrapping 146 base pairs of DNA in a left-handed super-helical turn sequentially to form chromosomal fiber. Two molecules of each of the four core histones (H2A, H2B, H3,, H4) form the octamer; formed of two H2A-H2B dimers, two H3-H4 dimers, forming two nearly symmetrical halves by tertiary structure. Over 80% of nucleosomes contain the linker Histone H1, derived from an intronless gene that interacts with linker DNA between nucleosomes, mediates compaction into higher order chromatin. Histones are subject to posttranslational modification by enzymes primarily on their N-terminal tails, but also in their globular domains. Such modifications include methylation, citrullination, acetylation, phosphorylation, sumoylation, ubiquitination, ADP-ribosylation. | |


-150x150.jpg)

There are no reviews yet.