Human, Canine, Mouse (-), and Rat (-) Anti-Microphthalmia Transcription Factor Antibody Product Attributes
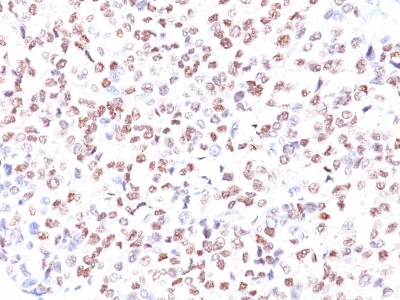
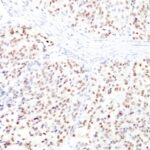
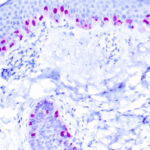
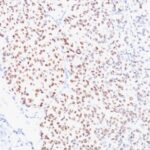
Microphthalmia Transcription Factor Previously Observed Antibody Staining Patterns
Observed Antibody Staining Data By Tissue Type:
Variations in Microphthalmia Transcription Factor antibody staining intensity in immunohistochemistry on tissue sections are present across different anatomical locations. An intense signal was observed in melanocytes in skin. More moderate antibody staining intensity was present in melanocytes in skin. Low, but measureable presence of Microphthalmia Transcription Factor could be seen in cells in the tubules in kidney, germinal center cells in the tonsil, glandular cells in the epididymis and stomach, hematopoietic cells in the bone marrow, lymphoid tissue in appendix, macrophages in lung, non-germinal center cells in the lymph node and tonsil and smooth muscle cells in the smooth muscle. We were unable to detect Microphthalmia Transcription Factor in other tissues. Disease states, inflammation, and other physiological changes can have a substantial impact on antibody staining patterns. These measurements were all taken in tissues deemed normal or from patients without known disease.
Observed Antibody Staining Data By Tissue Disease Status:
Tissues from cancer patients, for instance, have their own distinct pattern of Microphthalmia Transcription Factor expression as measured by anti-Microphthalmia Transcription Factor antibody immunohistochemical staining. The average level of expression by tumor is summarized in the table below. The variability row represents patient to patient variability in IHC staining.
| Sample Type | breast cancer | carcinoid | cervical cancer | colorectal cancer | endometrial cancer | glioma | head and neck cancer | liver cancer | lung cancer | lymphoma | melanoma | ovarian cancer | pancreatic cancer | prostate cancer | renal cancer | skin cancer | stomach cancer | testicular cancer | thyroid cancer | urothelial cancer |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Signal Intensity | – | – | – | – | – | – | – | – | – | – | +++ | – | – | – | – | – | – | – | + | – |
| MITF Variability | + | + | + | + | + | ++ | + | + | + | + | ++ | + | + | + | + | + | + | + | ++ | + |
| Microphthalmia Transcription Factor General Information | |
|---|---|
| Alternate Names | |
| Microphthalmia-associated transcription factor class E basic helix-loop-helix protein 32, bHLHe32, MITF | |
| Molecular Weight | |
| 52-56kDa (doublet) | |
| Chromosomal Location | |
| 3p14.1 | |
| Curated Database and Bioinformatic Data | |
| Gene Symbol | MITF |
| Entrez Gene ID | 4286 |
| Ensemble Gene ID | ENSG00000187098 |
| RefSeq Protein Accession(s) | XP_011532024, XP_016861935, NP_937801, NP_937820, XP_005264812, XP_016861936, NP_001341537, XP_005264811, XP_011532028, NP_001341534, XP_011532027, XP_016861937, NP_000239, NP_001171897, NP_001341533, NP_001341535, XP_011532025, NP_001171896, XP_016861933, XP_016861934, NP_001341536, NP_006713, XP_006713227, NP_937802, NP_937821 |
| RefSeq mRNA Accession(s) | NM_001354606, NM_198158, XM_017006447, NM_001354607, NM_006722, NM_198159, XM_006713164, XM_017006445, XM_017006446, NM_001184967, XM_011533722, NM_000248, NM_001354605, NM_198177, NM_001184968, XM_011533726, XM_017006448, XM_011533723, XM_011533725, XM_017006444, NM_001354608, XM_005264755, NM_001354604, NM_198178 |
| RefSeq Genomic Accession(s) | NC_000003, NC_018914, NG_011631, |
| UniProt ID(s) | A0A087WXU1, B4DNC7, O75030, Q8WYR3 |
| UniGene ID(s) | A0A087WXU1, B4DNC7, O75030, Q8WYR3 |
| HGNC ID(s) | 7105 |
| Cosmic ID(s) | MITF |
| KEGG Gene ID(s) | hsa:4286 |
| PharmGKB ID(s) | PA30823 |
| General Description of Microphthalmia Transcription Factor. | |
| MITF (microphthalmia transcription factor) is a basic helix-loop-helix-leucine-zipper (bHLH-Zip) transcription factor that regulates the development, survival of melanocytes, retinal pigment epithelium,, also is involved in transcription of pigmentation enzyme genes such as tyrosinase TRP1, TRP2. MITF has been shown to be phosphorylated by MAP kinase in response to c-kit activation, resulting in upregulation of MITF transcriptional activity. Mutations of the MITF gene are associated with the autosomal dominant hereditary deafness, pigmentation condition, Waardenburg Syndrome type 2A. Multiple isoforms of MITF exist, including MITF-A, MITF-B, MITF-C, MITF-H,, MITF-M, which differ in the amino-terminal domain, in their expression patterns. The MITF-M isoform is restricted to the melanocyte cell lineage. This MAb recognizes a nuclear protein, which is expressed in the majority of primary, metastatic epithelioid malignant melanomas as well as in normal melanocytes, benign nevi, dysplastic nevi. | |



There are no reviews yet.