Antibody (Suitable for clinical applications)
| Specification | Recommendation |
|---|---|
| Recommended Dilution (Conc) | 1:25-1:50 |
| Pretreatment | EDTA Buffer pH8.0 |
| Incubation Parameters | 60 min at Room Temperature |
Prior to use, inspect vial for the presence of any precipitate or other unusual physical properties. These can indicate that the antibody has degraded and is no longer suitable for patient samples. Please run positive and negative controls simultaneously with all patient samples to account and control for errors in laboratory procedure. Use of methods or materials not recommended by enQuire Bio including change to dilution range and detection system should be routinely validated by the user.
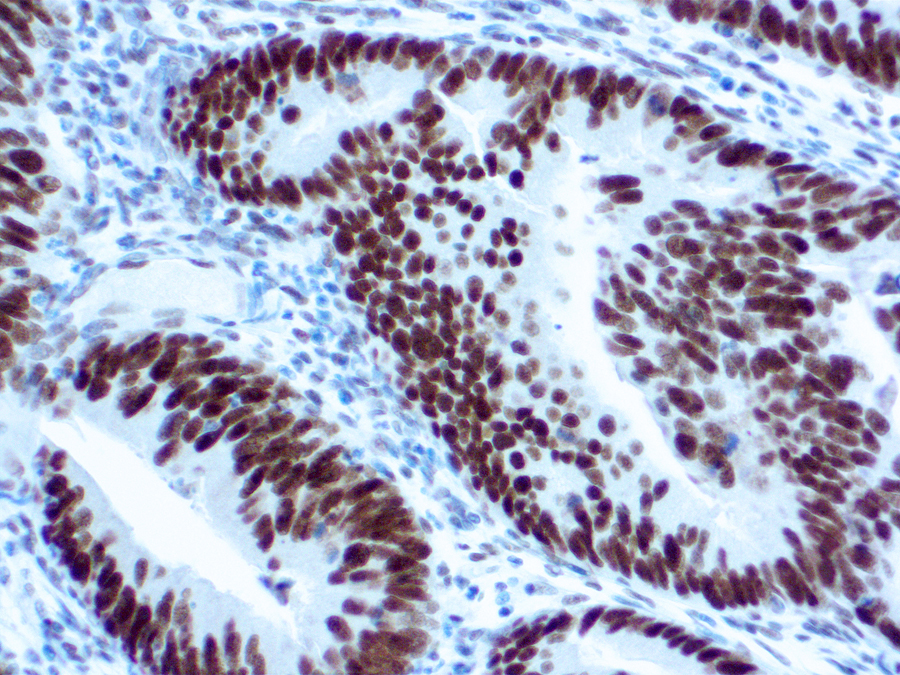
MLH-1 Information for Pathologists
Summary:
Mismatch repair gene. Mutations associated with Lynch syndrome (hereditary non-polyposis colon cancer) and some cases of sporadic colon cancer. Terminology Also called MutL homolog 1 colon cancer nonpolyposis type 2 (NCBI-Gene), hMLH1. Pathophysiology
Common Uses By Pathologists:
Use immunostaining for MLH1, MSH2, PMS2 and MSH6 to screen for Lynch syndrome. Biopsy samples may be as reliable as resections (Am J Surg Pathol 2011;35:447). Use of only PMS2 and MSH6 antibodies may be as effective as using all 4 antibodies (Am J Surg Pathol 2009;33:1639, Mod Pathol 2011;24:1004). Cases with normal staining due to somatic hypermethylation can occur (Am J Surg Pathol 2011;35:1902). Microscopic (histologic) images
| MLH-1 General Information | |
|---|---|
| Alternate Names | |
| Molecular Weight | |
| 84.6 kDa | |
| Chromosomal Location | |
| p22.2 [chr: 3] [chr_start: 36993332] [chr_end: 37050918] [strand: 1] | |
| Curated Database and Bioinformatic Data | |
| Gene Symbol | MLH1 |
| Entrez Gene ID | 4292 |
| RefSeq Protein Accession(s) | NP_000240; NP_001161089; NP_001161090; NP_001161091; XP_005265220; NP_001245200; XP_005265221; NP_001245203; NP_001245202 |
| RefSeq mRNA Accession(s) | XM_017006450; NM_001354621; NM_001354627; NM_001354628; NM_001354615; NM_001354619; NM_001354624; NM_001354625; NM_001167618; NM_001258273; NM_001258274; NM_001354617; NM_001354620; NM_001354622; NM_001354629; NM_001258271; NM_001354616; NM_001354626; NM_001167617; NM_001354618; NM_000249; NM_001354623; NM_001354630; XM_005265161; NM_001167619 |
| RefSeq Genomic Accession(s) | NG_007109; NC_000003 |
| UniProt ID(s) | P40692 |
| PharmGKB ID(s) | PA240 |
| KEGG Gene ID(s) | hsa:4292 |
| Associated Diseases (KEGG IDs) | Hereditary non-polyposis colorectal cancer 2 (HNPCC2) [MIM:609310]: An autosomal dominant disease associated with marked increase in cancer susceptibility. It is characterized by a familial predisposition to early-onset colorectal carcinoma (CRC) and extra-colonic tumors of the gastrointestinal, urological and female reproductive tracts. HNPCC is reported to be the most common form of inherited colorectal cancer in the Western world. Clinically, HNPCC is often divided into two subgroups. Type I is characterized by hereditary predisposition to colorectal cancer, a young age of onset, and carcinoma observed in the proximal colon. Type II is characterized by increased risk for cancers in certain tissues such as the uterus, ovary, breast, stomach, small intestine, skin, and larynx in addition to the colon. Diagnosis of classical HNPCC is based on the Amsterdam criteria: 3 or more relatives affected by colorectal cancer, one a first degree relative of the other two; 2 or more generation affected; 1 or more colorectal cancers presenting before 50 years of age; exclusion of hereditary polyposis syndromes. The term ‘suspected HNPCC’ or ‘incomplete HNPCC’ can be used to describe families who do not or only partially fulfill the Amsterdam criteria, but in whom a genetic basis for colon cancer is strongly suspected. {ECO:0000269|PubMed:10323887, ECO:0000269|PubMed:10375096, ECO:0000269|PubMed:10386556, ECO:0000269|PubMed:10413423, ECO:0000269|PubMed:10480359, ECO:0000269|PubMed:10598809, ECO:0000269|PubMed:10627141, ECO:0000269|PubMed:10660333, ECO:0000269|PubMed:10671064, ECO:0000269|PubMed:10713887, ECO:0000269|PubMed:10777691, ECO:0000269|PubMed:10882759, ECO:0000269|PubMed:11139242, ECO:0000269|PubMed:11427529, ECO:0000269|PubMed:11726306, ECO:0000269|PubMed:11748856, ECO:0000269|PubMed:11754112, ECO:0000269|PubMed:11781295, ECO:0000269|PubMed:11793442, ECO:0000269|PubMed:11839723, ECO:0000269|PubMed:11870161, ECO:0000269|PubMed:12095971, ECO:0000269|PubMed:12132870, ECO:0000269|PubMed:12200596, ECO:0000269|PubMed:12362047, ECO:0000269|PubMed:12373605, ECO:0000269|PubMed:12655562, ECO:0000269|PubMed:12658575, ECO:0000269|PubMed:14635101, ECO:0000269|PubMed:14961575, ECO:0000269|PubMed:15064764, ECO:0000269|PubMed:15139004, ECO:0000269|PubMed:15365995, ECO:0000269|PubMed:15365996, ECO:0000269|PubMed:16083711, ECO:0000269|PubMed:16451135, ECO:0000269|PubMed:17301300, ECO:0000269|PubMed:17510385, ECO:0000269|PubMed:18561205, ECO:0000269|PubMed:20020535, ECO:0000269|PubMed:21120944, ECO:0000269|PubMed:22753075, ECO:0000269|PubMed:7757073, ECO:0000269|PubMed:8566964, ECO:0000269|PubMed:8571956, ECO:0000269|PubMed:8797773, ECO:0000269|PubMed:8872463, ECO:0000269|PubMed:8993976, ECO:0000269|PubMed:9048925, ECO:0000269|PubMed:9067757, ECO:0000269|PubMed:9218993, ECO:0000269|PubMed:9272156, ECO:0000269|PubMed:9298827, ECO:0000269|PubMed:9311737, ECO:0000269|PubMed:9326924, ECO:0000269|PubMed:9399661, ECO:0000269|PubMed:9559627, ECO:0000269|PubMed:9718327, ECO:0000269|PubMed:9833759, ECO:0000269|PubMed:9927034, ECO:0000269|Ref.5}. The disease is caused by mutations affecting the gene represented in this entry.; Mismatch repair cancer syndrome (MMRCS) [MIM:276300]: An autosomal recessive, rare, childhood cancer predisposition syndrome encompassing a broad tumor spectrum. This includes hematological malignancies, central nervous system tumors, Lynch syndrome-associated malignancies such as colorectal tumors as well as multiple intestinal polyps, embryonic tumors and rhabdomyosarcoma. Multiple cafe-au-lait macules, a feature reminiscent of neurofibromatosis type 1, are often found as first manifestation of the underlying cancer. Areas of skin hypopigmentation have also been reported in MMRCS patients. {ECO:0000269|PubMed:11427529, ECO:0000269|PubMed:17440981, ECO:0000269|PubMed:7661930}. The disease is caused by mutations affecting the gene represented in this entry.; Muir-Torre syndrome (MRTES) [MIM:158320]: Rare autosomal dominant disorder characterized by sebaceous neoplasms and visceral malignancy. {ECO:0000269|PubMed:8751876}. The disease is caused by mutations affecting the gene represented in this entry.; Defects in MLH1 may contribute to lobular carcinoma in situ (LCIS), a non-invasive neoplastic disease of the breast.; Endometrial cancer (ENDMC) [MIM:608089]: A malignancy of endometrium, the mucous lining of the uterus. Most endometrial cancers are adenocarcinomas, cancers that begin in cells that make and release mucus and other fluids. Disease susceptibility is associated with variations affecting the gene represented in this entry.; Some epigenetic changes can be transmitted unchanged through the germline (termed ‘epigenetic inheritance’). Evidence that this mechanism occurs in humans is provided by the identification of individuals in whom 1 allele of the MLH1 gene is epigenetically silenced throughout the soma (implying a germline event). These individuals are affected by HNPCC but does not have identifiable mutations in MLH1, even though it is silenced, which demonstrates that an epimutation can phenocopy a genetic disease.; Colorectal cancer (CRC) [MIM:114500]: A complex disease characterized by malignant lesions arising from the inner wall of the large intestine (the colon) and the rectum. Genetic alterations are often associated with progression from premalignant lesion (adenoma) to invasive adenocarcinoma. Risk factors for cancer of the colon and rectum include colon polyps, long-standing ulcerative colitis, and genetic family history. {ECO:0000269|PubMed:10598809, ECO:0000269|PubMed:10882759, ECO:0000269|PubMed:12132870, ECO:0000269|PubMed:12655564, ECO:0000269|PubMed:14504054, ECO:0000269|PubMed:15184898, ECO:0000269|PubMed:18033691, ECO:0000269|PubMed:8872463, ECO:0000269|PubMed:9032648, ECO:0000269|PubMed:9087566, ECO:0000269|PubMed:9611074}. Disease susceptibility is associated with variations affecting the gene represented in this entry. |
| General Description of MLH-1 . | |
| The G168-15 antibody recognizes human and mouse MLH1 (80-85kDa). The repair of mismatch DNA is essential to maintaining the integrity of genetic information over time. An alteration of microsatellite repeats is the result of slippage owing to strand misalignment during DNA replication and is referred to as microsatellite instability (MSI). These defects in DNA repair pathways have been related to human carcinogenesis. The importance of mismatch repair genes became apparent with the identification of the genetic basis for hereditary nonpolyposis colon cancer (HNPC). MSH-2 is involved in the initial cognition of mismatch nucleotides during the replication mismatch repair process. It is thought that after MSH2 binds to a mismatched DNA duplex it is joined by a heterodimer of MLH1 and PMSH, which together help facilitate the later steps in mismatch repair. | |



Reviews
There are no reviews yet.