Human Anti-BCL10 Antibody Product Attributes
BCL10 Previously Observed Antibody Staining Patterns
Observed Subcellular, Organelle Specific Staining Data:
Anti-BCL10 antibody staining is expected to be primarily localized to the nucleoplasm.
Observed Antibody Staining Data By Tissue Type:
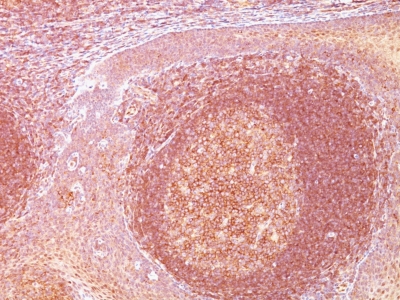
Variations in BCL10 antibody staining intensity in immunohistochemistry on tissue sections are present across different anatomical locations. Low, but measureable presence of BCL10 could be seen in cells in the seminiferous ducts in testis, exocrine glandular cells in the pancreas, follicle cells in the ovary, glandular cells in the epididymis, prostate, salivary gland, stomach and thyroid gland, melanocytes in skin, neuronal cells in the cerebral cortex, squamous epithelial cells in the cervix and uterine, esophagus, oral mucosa, tonsil and vagina. We were unable to detect BCL10 in other tissues. Disease states, inflammation, and other physiological changes can have a substantial impact on antibody staining patterns. These measurements were all taken in tissues deemed normal or from patients without known disease.
Observed Antibody Staining Data By Tissue Disease Status:
Tissues from cancer patients, for instance, have their own distinct pattern of BCL10 expression as measured by anti-BCL10 antibody immunohistochemical staining. The average level of expression by tumor is summarized in the table below. The variability row represents patient to patient variability in IHC staining.
| Sample Type | breast cancer | carcinoid | cervical cancer | colorectal cancer | endometrial cancer | glioma | head and neck cancer | liver cancer | lung cancer | lymphoma | melanoma | ovarian cancer | pancreatic cancer | prostate cancer | renal cancer | skin cancer | stomach cancer | testicular cancer | thyroid cancer | urothelial cancer |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Signal Intensity | + | – | + | ++ | + | – | + | – | + | +++ | + | + | + | + | – | ++ | ++ | – | + | + |
| BCL10 Variability | ++ | ++ | ++ | ++ | ++ | + | + | ++ | +++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| BCL10 General Information | |
|---|---|
| Alternate Names | |
| B-cell lymphoma 10, B-cell leukemia 10, BCL10, BCL-10 | |
| Molecular Weight | |
| 33kDa | |
| Chromosomal Location | |
| 1p22.3 | |
| Curated Database and Bioinformatic Data | |
| Gene Symbol | BCL10 |
| Entrez Gene ID | 8915 |
| Ensemble Gene ID | ENSG00000142867 |
| RefSeq Protein Accession(s) | XP_011540700, NP_001307644, NP_003912, XP_011540701, XP_011540699 |
| RefSeq mRNA Accession(s) | XM_011542397, XM_011542398, NM_003921, NM_001320715 |
| RefSeq Genomic Accession(s) | NG_012216, NC_000001, NC_018912 |
| UniProt ID(s) | O95999, A2TDT2 |
| UniGene ID(s) | O95999, A2TDT2 |
| HGNC ID(s) | 989 |
| Cosmic ID(s) | BCL10 |
| KEGG Gene ID(s) | hsa:8915 |
| PharmGKB ID(s) | PA25299 |
| General Description of BCL10. | |
| BCL10, with an N-terminal caspase recruitment domain (CARD), is found in a number of apoptotic regulatory molecules. It was identified through its direct involvement in t(1;14) of mucosa-associated lymphoid tissue (MALT) lymphoma. Expression of BCL10 was shown to induce NF??B activation in a NIK-dependent pathway. This MAb labels subpopulations of normal B, T cells, is a useful tool for the sub-classification of lymphomas. In MALT lymphomas with the t(1;14) translocation, while 55% of MALT lymphomas lacking this translocation exhibited the same labeling pattern, although at a much lower level. | |




There are no reviews yet.