Human Anti-EGFR Antibody Product Attributes
EGFR Previously Observed Antibody Staining Patterns
Observed Subcellular, Organelle Specific Staining Data:
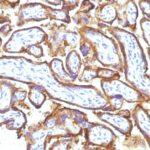
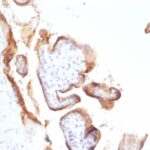
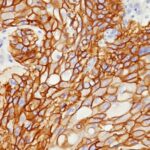
Variations in EGFR antibody staining intensity in immunohistochemistry on tissue sections are present across different anatomical locations. An intense signal was observed in trophoblastic cells in the placenta. More moderate antibody staining intensity was present in trophoblastic cells in the placenta. Low, but measureable presence of EGFR could be seen inhepatocytes in liver, Leydig cells in the testis, melanocytes in skin, myocytes in skeletal muscle, squamous epithelial cells in the tonsil and vagina and urothelial cells in the urinary bladder. We were unable to detect EGFR in other tissues. Disease states, inflammation, and other physiological changes can have a substantial impact on antibody staining patterns. These measurements were all taken in tissues deemed normal or from patients without known disease.
Observed Antibody Staining Data By Tissue Type:
Tissues from cancer patients, for instance, have their own distinct pattern of EGFR expression as measured by anti-EGFR antibody immunohistochemical staining. The average level of expression by tumor is summarized in the table below. The variability row represents patient to patient variability in IHC staining.
| Sample Type | breast cancer | carcinoid | cervical cancer | colorectal cancer | endometrial cancer | glioma | head and neck cancer | liver cancer | lung cancer | lymphoma | melanoma | ovarian cancer | pancreatic cancer | prostate cancer | renal cancer | skin cancer | stomach cancer | testicular cancer | thyroid cancer | urothelial cancer |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Signal Intensity | – | – | – | – | – | + | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| EGFR Variability | + | + | + | + | + | ++ | + | ++ | + | + | + | + | + | + | + | + | + | + | + | + |
| EGFR General Information | |
|---|---|
| Alternate Names | |
| EGFR, ErbB-1, HER1, ErbB1 | |
| Molecular Weight | |
| 170kDa (wild type) and 145kDa (vIII variant) | |
| Chromosomal Location | |
| 7p11.2 | |
| Curated Database and Bioinformatic Data | |
| Gene Symbol | EGFR |
| Entrez Gene ID | 1956 |
| Ensemble Gene ID | ENSG00000146648 |
| RefSeq Protein Accession(s) | NP_001333827, NP_958439, NP_001333829, NP_958440, NP_001333826, NP_958441, NP_005219, NP_001333870, NP_001333828 |
| RefSeq mRNA Accession(s) | NM_201282, NM_001346897, NM_201283, NM_001346898, NM_001346941 NM_201284, NM_001346900, NM_001346899, NM_005228 |
| RefSeq Genomic Accession(s) | NC_018918, NG_007726, NC_000007 |
| UniProt ID(s) | Q504U8, B7Z2I3, E9PFD7, P00533, F2YGG7, E7BSV0 |
| UniGene ID(s) | Q504U8, B7Z2I3, E9PFD7, P00533, F2YGG7, E7BSV0 |
| HGNC ID(s) | 3236 |
| Cosmic ID(s) | EGFR |
| KEGG Gene ID(s) | hsa:1956 |
| PharmGKB ID(s) | PA7360 |
| General Description of EGFR. | |
| This MAb reacts with a cytoplasmic domain of EGFR. EGFR is a type I receptor tyrosine kinase with sequence homology to erbB-1, -2, -3 -4 or HER-1, -2, -3 -4. It binds to Epidermal Growth Factor (EGF), Transforming Growth Factor-a (TGF-a), Heparin-binding EGF (HB-EGF), amphiregulin, betacellulin & epiregulin. | |

![Analysis of Mass Spec data (dashed-line) of fractions stained with EGFR MS-QAVA™ monoclonal antibody [Clone: H9B4] (solid-line), reveals that less than 13.1% of signal is attributable to non-specific binding of anti-EGFR [Clone H9B4] to targets other than EGFR protein. Even frequently cited antibodies have much greater non-specific interactions, averaging over 30%. Data in image is from analysis in Jurkat, U202 and HeLa cells.](https://cdn-enquirebio.pressidium.com/wp-content/uploads/2017/10/enQuire-Bio-1956-MSM3-P1-anti-EGFR-antibody.png)


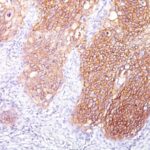
![Staining with mouse monoclonal EGFR [Clone 31G7] antibody in formalin-fixed paraffin-embedded human placenta.](https://enquirebio.com/wp-content/uploads/2019/01/EGFR IHC human Placenta (31G7)-150x150.jpg)
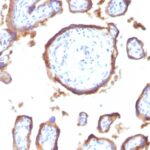
-150x150.jpg)

There are no reviews yet.