Human, Mouse, Rat, and chicken Anti-NKX2.2 Antibody Product Attributes
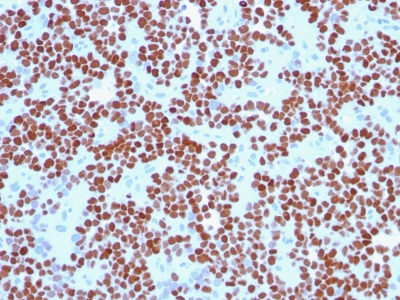
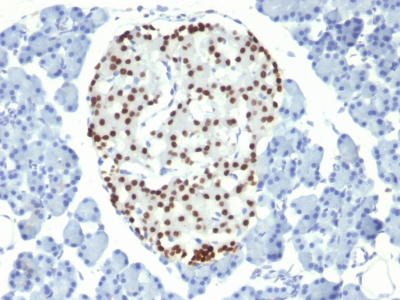
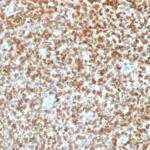
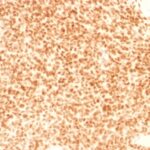
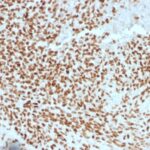
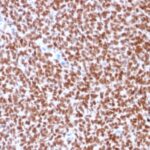
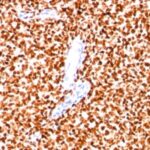
NKX2.2 Previously Observed Antibody Staining Patterns
Observed Antibody Staining Data By Tissue Type:
Variations in NKX2.2 antibody staining intensity in immunohistochemistry on tissue sections are present across different anatomical locations. Low, but measureable presence of NKX2.2 could be seen in cells in the molecular layer in cerebellum, glandular cells in the duodenum and epididymis, neuronal cells in the hippocampus, cells in the tubules in kidney, exocrine glandular cells in the pancreas, islets of Langerhans in pancreas, glandular cells in the seminal vesicle, epidermal cells in the skin, glandular cells in the small intestine and stomach, cells in the seminiferous ducts in testis, Leydig cells in the testis, non-germinal center cells in the tonsil and squamous epithelial cells in the tonsil. We were unable to detect NKX2.2 in other tissues. Disease states, inflammation, and other physiological changes can have a substantial impact on antibody staining patterns. These measurements were all taken in tissues deemed normal or from patients without known disease.
Observed Antibody Staining Data By Tissue Disease Status:
Tissues from cancer patients, for instance, have their own distinct pattern of NKX2.2 expression as measured by anti-NKX2.2 antibody immunohistochemical staining. The average level of expression by tumor is summarized in the table below. The variability row represents patient to patient variability in IHC staining.
| Sample Type | breast cancer | carcinoid | cervical cancer | colorectal cancer | endometrial cancer | glioma | head and neck cancer | liver cancer | lung cancer | lymphoma | melanoma | ovarian cancer | pancreatic cancer | prostate cancer | renal cancer | skin cancer | stomach cancer | testicular cancer | thyroid cancer | urothelial cancer |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Signal Intensity | ++ | + | – | + | + | + | – | – | – | – | – | – | – | ++ | + | – | + | – | – | – |
| NKX2-2 Variability | ++ | ++ | + | ++ | ++ | ++ | ++ | + | + | + | ++ | ++ | ++ | + | ++ | + | ++ | ++ | + | ++ |
| NKX2.2 General Information | |
|---|---|
| Alternate Names | |
| Homeobox protein Nkx-2, NKX2-2 | |
| Molecular Weight | |
| 40-50kDa | |
| Chromosomal Location | |
| 20p11.22 | |
| Curated Database and Bioinformatic Data | |
| Gene Symbol | NKX2-2 |
| Entrez Gene ID | 4821 |
| Ensemble Gene ID | ENSG00000125820 |
| RefSeq Protein Accession(s) | NP_002500, XP_006723629 |
| RefSeq mRNA Accession(s) | XM_006723566, NM_002509, |
| RefSeq Genomic Accession(s) | NC_000020, NG_042186, NC_018931 |
| UniProt ID(s) | O95096 |
| UniGene ID(s) | O95096 |
| HGNC ID(s) | 7835 |
| Cosmic ID(s) | NKX2-2 |
| KEGG Gene ID(s) | hsa:4821 |
| PharmGKB ID(s) | PA31642 |
| General Description of NKX2.2. | |
| Expression of NKX2.2 has been found in neuroendocrine tumors of the gut, making it a potential marker for the study of gastrointestinal neuroendocrine tumors. More recently, NKX2.2 protein was identified as a target of EWS-FLI-1, the fusion protein specific to Ewing sarcoma,, was shown to be differentially upregulated in Ewing sarcoma on the basis of array-based gene expression analysis. It acts as a valuable marker for Ewing sarcoma, with a sensitivity of 93%, a specificity of 89%,, aids in the differential diagnosis of small round cell tumors. | |




There are no reviews yet.