Human CETN2 / Centrin 2 Recombinant Protein Product Attributes
Product Type: Recombinant Protein
Recombinant CETN2 / Centrin 2 based upon sequence from Human
Host: QP5820 protein expressed in E. coli.
Tag: His-SUMO
Protein Construction: A DNA sequence encoding the Homo sapiens (Human) CETN2 / Centrin 2, was expressed in the hosts and tags indicated. Please select your host/tag option, above.
Recommended Applications: Immunogen, Protein Standard, Cell culture, or Other Cell Biology Applications.
Application Notes: Please contact us for application specific information for QP5820.
Bioactivity Data: Untested
Full Length? Full Length
Expression Region: Met1 – Tyr172
Amino Acid Sequence: MASNFKKANM ASSSQRKRMS PKPELTEEQK QEIREAFDLF DADGTGTIDV KELKVAMRAL GFEPKKEEIK KMISEIDKEG TGKMNFGDFL TVMTQKMSEK DTKEEILKAF KLFDDDETGK ISFKNLKRVA KELGENLTDE ELQEMIDEAD RDGDGEVSEQ EFLRIMKKTS LY
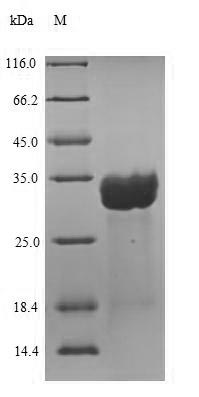
Purity: Greater than 90% as determined by SDS-PAGE.
Reconstitution Instructions: Concentrated protein in liquid format. Reconstitution is not necessary.
Concentration of Human CETN2 / Centrin 2 Protein:
Endotoxin Levels: Not determined.
Buffer: Tris-based buffer, 50% glycerol
Storage Conditions: Store at -20C to -80C.
| Recombinant Human CETN2 / Centrin 2 Protein General Information | |
|---|---|
| Alternate Names | |
| CETN2, Centrin 2, CEN2, CALT | |
| Curated Database and Bioinformatic Data | |
| Gene Symbol | CETN2 |
| Entrez Gene ID | 1069 |
| Ensemble Gene ID | ENSG00000147400 |
| RefSeq Protein Accession(s) | NP_004335.1 |
| RefSeq mRNA Accession(s) | NM_004344.1 |
| UniProt ID(s) | P41208 |
| UniGene ID(s) | Hs.82794 |
| HGNC ID(s) | HGNC:1867 |
| COSMIC ID Link(s) | CETN2 |
| KEGG Gene ID(s) | hsa:1069 |
| PharmGKB ID(s) | PA26420 |
| General Description of Recombinant Human CETN2 / Centrin 2 Protein. | |
| Plays a fundamental role in microtubule organizing center structure and function. Required for centriole duplication and correct spindle formation. Has a role in regulating cytokinesis and genome stability via cooperation with CALM1 and CCP110.Involved in global genome nucleotide excision repair (GG-NER) by acting as component of the XPC complex. Cooperatively with RAD23B appears to stabilize XPC. In vitro, stimulates DNA binding of the XPC:RAD23B dimer.The XPC complex is proposed to represent the first factor bound at the sites of DNA damage and together with other core recognition factors, XPA, RPA and the TFIIH complex, is part of the pre-incision (or initial recognition) complex. The XPC complex recognizes a wide spectrum of damaged DNA characterized by distortions of the DNA helix such as single-stranded loops, mismatched bubbles or single-stranded overhangs. The orientation of XPC complex binding appears to be crucial for inducing a productive NER. XPC complex is proposed to recognize and to interact with unpaired bases on the undamaged DNA strand which is followed by recruitment of the TFIIH complex and subsequent scanning for lesions in the opposite strand in a 5′-to-3′ direction by the NER machinery. Cyclobutane pyrimidine dimers (CPDs) which are formed upon UV-induced DNA damage esacpe detection by the XPC complex due to a low degree of structural perurbation. Instead they are detected by the UV-DDB complex which in turn recruits and cooperates with the XPC complex in the respective DNA repair.Component of the TREX-2 complex (transcription and export complex 2), composed of at least ENY2, GANP, PCID2, DSS1, and either centrin CETN2 or CETN3 . The TREX-2 complex functions in docking export-competent ribonucleoprotein particles (mRNPs) to the nuclear entrance of the nuclear pore complex (nuclear basket). TREX-2 participates in mRNA export and accurate chromatin positioning in the nucleus by tethering genes to the nuclear periphery . | |
Limitations and Performance Guarantee
This is a life science research product (for Research Use Only). This product is guaranteed to work for a period of two years when stored at -70C or colder, and one year when aliquoted and stored at -20C.



There are no reviews yet.