Human HRV-1A Recombinant Protein Product Attributes
Product Type: Recombinant Protein
Recombinant HRV-1A based upon sequence from: Human
Host: QP9561 protein expressed in Yeast.
Tag: His
Protein Construction: A DNA sequence encoding the Homo sapiens (Human) HRV-1A, was expressed in the hosts and tags indicated. Please select your host/tag option, above.
Application Notes: Please contact us for application specific information for QP9561.
Bioactivity Data: Untested
Full Length? Partial (see sequence information for more details).
Expression Region: Asn571 – Ala857
Amino Acid Sequence: NPVENYIDEV LNEVLVVPNI KESHHTTSNS APLLDAAETG HTSNVQPEDA IETRYVITSQ TRDEMSIESF LGRSGCVHIS RIKVDYTDYN GQDINFTKWK ITLQEMAQIR RKFELFTYVR FDSEITLVPC IAGRGDDIGH IVMQYMYVPP GAPIPSKRND FSWQSGTNMS IFWQHGQPFP RFSLPFLSIA SAYYMFYDGY DGDNTSSKYG SVVTNDMGTI CSRIVTEKQK HSVVITTHIY HKAKHTKAWC PRPPRAVPYT HSHVTNYMPE TGDVTTAIVR RNTITTA
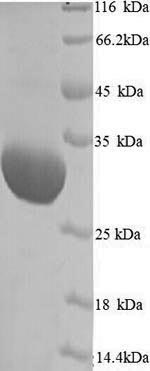
Purity: Greater than 90% as determined by SDS-PAGE.
Reconstitution Instructions:
Concentration of Human HRV-1A Protein:
Endotoxin Levels: Not determined.
Buffer: Tris-based buffer, 50% glycerol
Storage Conditions: Store at -20C to -80C.
| Recombinant Human HRV-1A Protein General Information | |
|---|---|
| Curated Database and Bioinformatic Data | |
| UniProt ID(s) | P23008 |
| General Description of Recombinant Human HRV-1A Protein. | |
| Capsid protein VP1: Forms an icosahedral capsid of pseudo T=3 symmetry with capsid proteins VP2 and VP3. The capsid is 300 Angstroms in diameter, composed of 60 copies of each capsid protein and enclosing the viral positive strand RNA genome. Capsid protein VP1 mainly forms the vertices of the capsid. Capsid protein VP1 interacts with host cell receptor to provide virion attachment to target host cells. This attachment induces virion internalization. Tyrosine kinases are probably involved in the entry process. After binding to its receptor, the capsid undergoes conformational changes. Capsid protein VP1 N-terminus (that contains an amphipathic alpha-helix) and capsid protein VP4 are externalized. Together, they shape a pore in the host mbrane through which viral genome is translocated to host cell cytoplasm. After genome has been released, the channel shrinks .Capsid protein VP2: Forms an icosahedral capsid of pseudo T=3 symmetry with capsid proteins VP2 and VP3. The capsid is 300 Angstroms in diameter, composed of 60 copies of each capsid protein and enclosing the viral positive strand RNA genome .Capsid protein VP3: Forms an icosahedral capsid of pseudo T=3 symmetry with capsid proteins VP2 and VP3. The capsid is 300 Angstroms in diameter, composed of 60 copies of each capsid protein and enclosing the viral positive strand RNA genome .Capsid protein VP4: Lies on the inner surface of the capsid shell. After binding to the host receptor, the capsid undergoes conformational changes. Capsid protein VP4 is released, Capsid protein VP1 N-terminus is externalized, and together, they shape a pore in the host mbrane through which the viral genome is translocated into the host cell cytoplasm. After genome has been released, the channel shrinks .Capsid protein VP0: Component of immature procapsids, which is cleaved into capsid proteins VP4 and VP2 after maturation. Allows the capsid to rain inactive before the maturation step .Protein 2A: Cysteine protease that cleaves viral polyprotein and specific host proteins. It is responsible for the cleavage between the P1 and P2 regions, first cleavage occurring in the polyprotein. Cleaves also the host translation initiation factor EIF4G1, in order to shut down the capped cellular mRNA translation. Inhibits the host nucleus-cytoplasm protein and RNA trafficking by cleaving host mbers of the nuclear pores .Protein 2B: Plays an essential role in the virus replication cycle by acting as a viroporin. Creates a pore in the host reticulum endoplasmic and as a consequence releases Ca2+ in the cytoplasm of infected cell. In turn, high levels of cyctoplasmic calcium may trigger mbrane trafficking and transport of viral ER-associated proteins to viroplasms, sites of viral genome replication .Protein 2C: Induces and associates with structural rearrangents of intracellular mbranes. Displays RNA-binding, nucleotide binding and NTPase activities. May play a role in virion morphogenesis and viral RNA encapsidation by interacting with the capsid protein VP3 .Protein 3AB: Localizes the viral replication complex to the surface of mbranous vesicles. Together with protein 3CD binds the Cis-Active RNA Elent (CRE) which is involved in RNA synthesis initiation. Acts as a cofactor to stimulate the activity of 3D polymerase, maybe through a nucleid acid chaperone activity .Protein 3A: Localizes the viral replication complex to the surface of mbranous vesicles. It inhibits host cell endoplasmic reticulum-to-Golgi apparatus transport and causes the dissassbly of the Golgi complex, possibly through GBF1 interaction. This would result in depletion of MHC, trail receptors and IFN receptors at the host cell surface .Viral protein genome-linked: acts as a primer for viral RNA replication and rains covalently bound to viral genomic RNA. VPg is uridylylated prior to priming replication into VPg-pUpU. The oriI viral genomic sequence may act as a tplate for this. The VPg-pUpU is then used as primer on the genomic RNA poly(A) by the RNA-dependent RNA polymerase to replicate the viral genome. VPg may be roved in the cytoplasm by an unknown enzyme termed “unlinkase”. VPg is not cleaved off virion genomes because replicated genomic RNA are encapsidated at the site of replication .Protein 3CD: Is involved in the viral replication complex and viral polypeptide maturation. It exhibits protease activity with a specificity and catalytic efficiency that is different from protease 3C. Protein 3CD lacks polymerase activity. The 3C domain in the context of protein 3CD may have an RNA binding activity .Protease 3C: cleaves host DDX58/RIG-I and thus contributes to the inhibition of type I interferon production. Cleaves also host PABPC1 .RNA-directed RNA polymerase: Replicates the viral genomic RNA on the surface of intracellular mbranes. May form linear arrays of subunits that propagate along a strong head-to-tail interaction called interface-I. Covalently attaches UMP to a tyrosine of VPg, which is used to prime RNA synthesis. The positive stranded RNA genome is first replicated at virus induced mbranous vesicles, creating a dsRNA genomic replication form. This dsRNA is then used as tplate to synthesize positive stranded RNA genomes. ss+RNA genomes are either translated, replicated or encapsidated .. | |
Limitations and Performance Guarantee
This is a life science research product (for Research Use Only). This product is guaranteed to work for a period of two years when stored at -70C or colder, and one year when aliquoted and stored at -20C.



There are no reviews yet.