Human Anti-CD3 Antibody Product Attributes
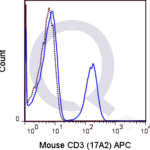
CD3 Previously Observed Antibody Staining Patterns
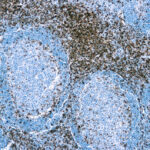
Observed Antibody Staining Data By Tissue Type:
Variations in CD3 antibody staining intensity in immunohistochemistry on tissue sections are present across different anatomical locations. An intense signal was observed in cells in the red pulp in spleen and non-germinal center cells in the lymph node and tonsil. More moderate antibody staining intensity was present in cells in the red pulp in spleen and non-germinal center cells in the lymph node and tonsil. Low, but measureable presence of CD3 could be seen in. We were unable to detect CD3 in other tissues. Disease states, inflammation, and other physiological changes can have a substantial impact on antibody staining patterns. These measurements were all taken in tissues deemed normal or from patients without known disease.
Observed Antibody Staining Data By Tissue Disease Status:
Tissues from cancer patients, for instance, have their own distinct pattern of CD3 expression as measured by anti-CD3 antibody immunohistochemical staining. The average level of expression by tumor is summarized in the table below. The variability row represents patient to patient variability in IHC staining.
| Sample Type | breast cancer | carcinoid | cervical cancer | colorectal cancer | endometrial cancer | glioma | head and neck cancer | liver cancer | lung cancer | lymphoma | melanoma | ovarian cancer | pancreatic cancer | prostate cancer | renal cancer | skin cancer | stomach cancer | testicular cancer | thyroid cancer | urothelial cancer |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Signal Intensity | – | – | – | – | – | – | – | – | – | ++ | – | – | – | – | – | – | – | – | – | – |
| CD3D Variability | + | + | + | + | + | ++ | + | ++ | + | ++ | + | + | ++ | + | + | + | + | ++ | ++ | + |
| CD3 General Information | |
|---|---|
| Alternate Names | |
| Leu-4, T3 | |
| Curated Database and Bioinformatic Data | |
| Gene Symbol | CD3D |
| Entrez Gene ID | 915 |
| Ensemble Gene ID | ENSG00000167286 |
| RefSeq Protein Accession(s) | XP_016874032, NP_000723, NP_001035741 |
| RefSeq mRNA Accession(s) | NM_000732, NM_001040651 |
| RefSeq Genomic Accession(s) | NC_000011, NG_009891, NC_018922 |
| UniProt ID(s) | B0YIY4, P04234 |
| UniGene ID(s) | B0YIY4, P04234 |
| HGNC ID(s) | 1673 |
| Cosmic ID(s) | CD3D |
| KEGG Gene ID(s) | hsa:915 |
| PharmGKB ID(s) | PA26215 |
| General Description of CD3. | |
| The UCHT1 antibody is specific for human CD3e, also known as CD3 epsilon, a 20 kDa subunit of the T cell receptor complex, along with CD3 gamma and CD3 delta. These integral membrane protein chains assemble with additional chains of the T cell receptor (TCR), as well as CD3 zeta chain, to form the T cell receptor CD3 complex. Together with co-receptors CD4 or CD8, the complex serves to recognize antigens bound to MHC molecules on antigen-presenting cells. These interactions promote T cell receptor signaling (T cell activation), inducing cell proliferation, differentiation, production of cytokines or activation-induced cell death. CD3 is differentially expressed during thymocyte-to-T cell development and on all mature T cells.The UCHT1 antibody is a widely used phenotypic marker for human T cells. In addition, binding/cross-linking of UCHT1 antibody to CD3e can induce cell activation (use format suitable for functional assays). A recent publication of the crystal structure of a CD3e- antibody complex provides insight as to the action of commonly used agonist antibodies, as well as specific epitope-binding data for the human CD3 antibodies UCHT1 and OKT3 (Fernandes, R.A. et al. 2012. J. Biol. Chem. 287: 13324-13335).UCHT1 antibody reacts with both surface-expressed and intracellular CD3e protein, in contrast to an alternative human CD3 clone, Hit3a, which will stain only the extracellular (membrane-expressed) CD3e protein. Also, the UCHT1 antibody is reported to be cross-reactive with chimpanzee and has been used for phenotypic analysis of expression by flow cytometry; however the antibody is reported to be unsuitable for induction of T cell activation in this species (Bibollet-Ruche et al. 2009. J. Virol. 82: 10271-10278). | |

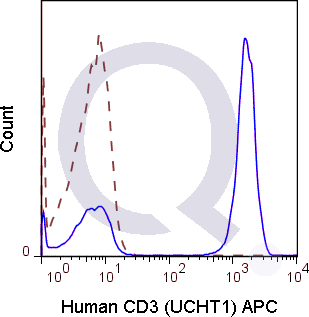

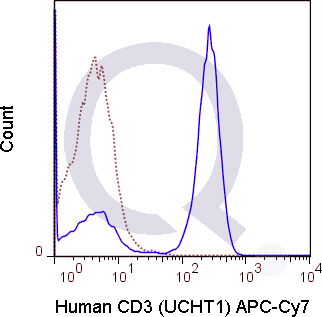

![Anti-CD3 Antibody [UCHT1] - Image 5](https://cdn-enquirebio.pressidium.com/wp-content/uploads/2017/10/enQuire-Bio-QAB6-F-100Tests-anti-CD3-antibody-10.png)
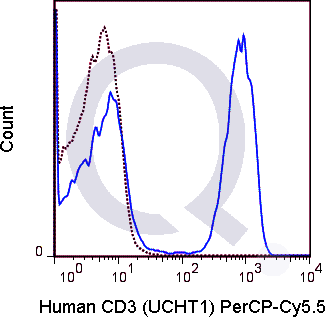
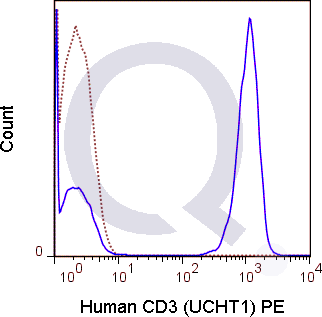
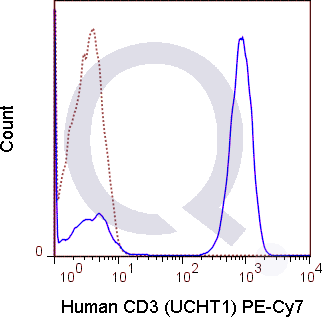
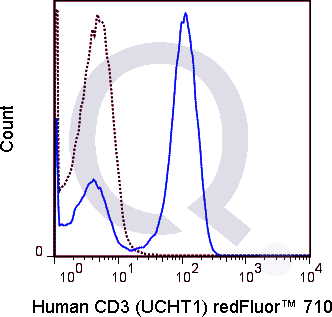

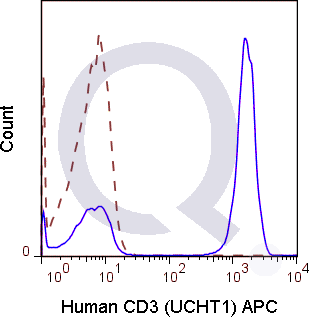
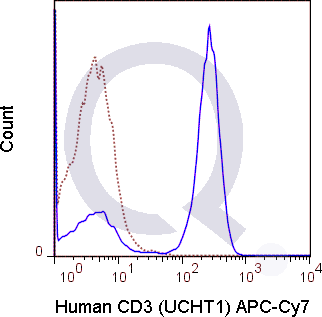
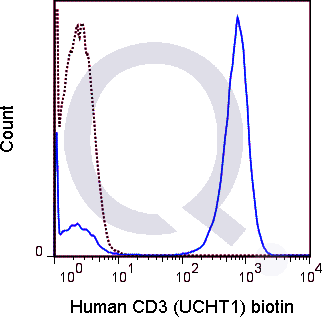
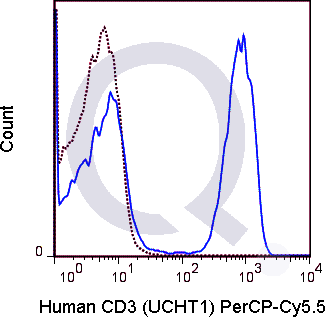
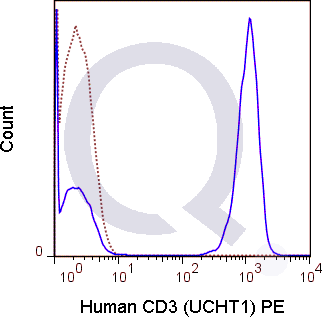
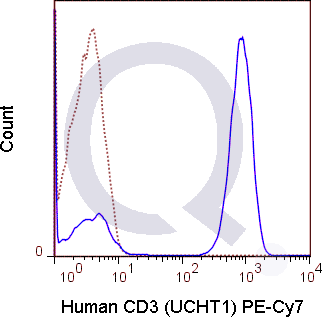
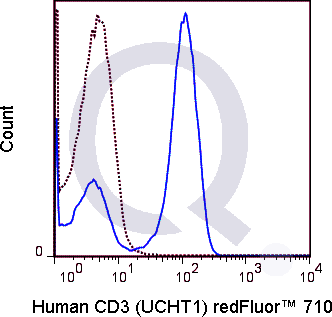
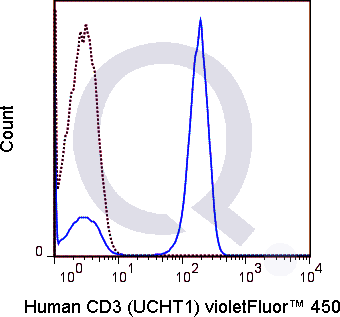
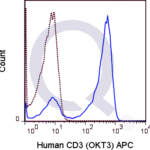
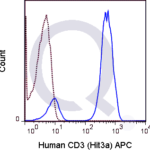



There are no reviews yet.